μBrain
a 3D volumetric reconstruction of the mid-gestation fetal brain
About μBrainLearn more about the project
View the Project on GitHub garedaba/micro-brain
Download the Data from Zenodo3D volume, surfaces and microarray data
Important information
μBrain is a under construction and subject to revision.
Built upon existing open-source data, the μBrain atlas is a new and freely-available 3D volumetric model of the mid-fetal brain.
Using serial Nissl-stained and anatomically-labelled sections of the prenatal brain (Ding et al., 2022), we performed automated image repair using a generative neural network model before aligning all sections into a common anatomical space, resulting in a 3D volume at 150μm voxel resolution.
The μBrain volume is accompanied by a set of brain tissue labels (n=20) and surface models of the cortical plate surface. The cortical surface is further parcellated into a set of cytoarchitecturally-defined labels (n=29). To faciliate integration with fetal and neonatal neuroimaging studies, the cortical surfaces and parcellations have been aligned to the fetal surface template of the Developing Human Connectome Project.
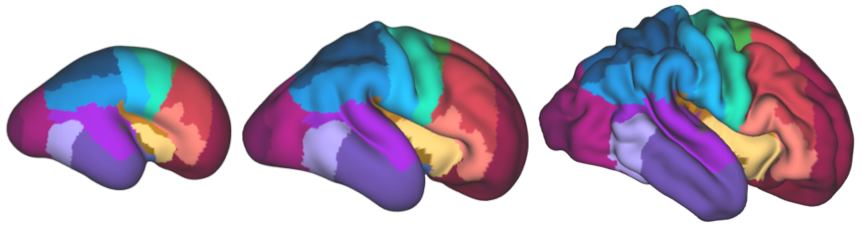
In addition, cortical areas are matched to normalised gene expression data from corresponding laser microdissection microarrays across multiple tissue zones in three additional prenatal specimens (Miller et al. 2014)
μBrain is a three-dimensional, high resolution histological atlas of the human fetal brain, coupled with bulk tissue microarray data, sampled across 29 cortical regions and 5 transient tissue zones. It provides a 3D anatomical coordinate space to facilitate integrated imaging-transcriptomic analyses of the developing brain.
Data descriptors
Volumetric data
μBrain-volume.nii.gz
a 3D reconstruction of the cerebral hemisphere at 150μm resolution
μBrain-atlas-labels.nii.gz
corresponding brain tissue labels (n=20)
brain-tissue-labels.txt
look-up table for brain tissue labels
Surface data
μBrain.R.inner/outer.surf.gii
inner and outer cortical surfaces of the μBrain volume
μBrain.cortical-atlas.fetal36w-template.label.gii
μBrain cortical atlas projected onto the dHCP template surface
cortical_labels.txt
look-up table for cortical atlas labels
Microarray data
μBrain-processed-lmd-data.csv
LMD microarray data aligned to the μBrain cortical atlas in long format. Each row contains normalised expression for a single observation. Each observation is identified by tissue (cortical plate, subplate, etc), cortical region in μBrain, specimen and gene.
Data resources & references
Ding, S.-L. et al. Cellular resolution anatomical and molecular atlases for prenatal human brains. J. Comp. Neurol. 530, 6–503 (2022)
Miller, J. A. et al. Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014)
Project funding
